Update: I just realized that wordpress truncated my DNA sequences at the bottom of this post. If you would like to get the full sequences, you can get them by “view source” in your browser. If that doesn’t work, let me know.
This post is the first part of a three-post series aimed at clearing up the misinformation written by the Discovery Institute’s Casey Luskin. In his recent posts, he tries to persuade his readers that the idea of a tree of life (TOL) and the very idea of phylogenetic trees is erroneous and not evidence of common descent. These trees are created by looking at genetic similarities between organisms to arrange them in terms of relatedness and common ancestry. In my series of posts, I will expose the weaknesses in the arguments put forth by Luskin.
Part 1 – Why we know the tree is real
In the first part of my three-part series on the realities of the TOL, I will cover some of the reasons why we know it is real and why there are difficulties in creating a single tree that encompasses all organisms. This post doesn’t directly address Luskin’s concerns, but is more of an overview. Point by point analysis will come later.
First, I should point out that when I write “tree,” I am not referring to a perfect tree with a single trunk at the base and straightforward branching all the way to the top. I am referring to a tree that does have plenty of instances with a typical branching pattern, but also one that has tangled, intertwining branches. This is a widely held view among scientists in the field and was written about nearly 10 years ago in Scientific American by Dr. Ford Doolittle.
Without further ado, lets look at why the idea of a phylogenetic tree of life is valid:
Trees can be created that match observations – This simple fact alone should be enough to show the validity of phylogenetic trees. If common descent wasn’t true, then any phylogenetic tree should be meaningless, but they generally follow the organization of standard taxonomy that we have known for years. Furthermore, these trees are very similar to each other when different genes or combination of genes are used.
Other new lines of evidence correlate with trees base on gene similarities – One good way to see if a scientific idea is valid is to see how well it correlates with other data. The phylogenetic TOL correlates with taxonomic observations, but what about other lines of evidence for common descent? For this post, I decided to perform a little experiment. It has previously been shown (Lebedev et al. 2000) that the introduction of endogenous retroviruses (ERVs) into genomes can be used to created phylogenetic trees. For a discussion of this topic, see here. This technique was used in primates to create the tree shown below on the left. I decided to see if a tree created by using the similarities in the gene for cytochrome B would yield the same tree (Details on exactly how I did this are at the bottom of this post). As shown below, it did match exactly.
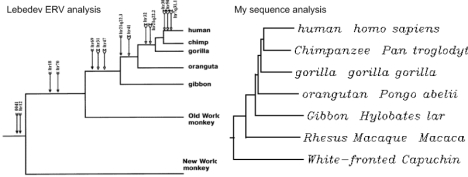
Comparison of trees made by ERV or sequence comparison
Two different techniques yielded the exact same tree. Both trees created here match to a tree that is created by looking at physical similarities and differences. This one example should be enough to show the validity of phylogenetic trees, but you know nothing would be sufficient to convince the Discovery Institute.
So I have shown a couple reasons why I think phylogenetic trees are valid. Keep these in mind when you read about the supposed failures of phylogenetic trees from the ID proponents. Next, I am going to describes some of the difficulties in creating a complete TOL. Remember, these issues do not mean that common descent is wrong, it just means that our ability to decipher the tree of life is limited. A weakness in data is not a weakness in a theory.
Difficulties in creating a tree of life
There are many problems with creating a complete tree of life that takes into account every piece of data available. Think about all the information and computation required for the creation of an all-encompassing phylogenetic tree. In addition, a TOL has to determine the relatedness of organisms that might not have a common ancestor for over half a billion years. Below is an overview of some other problems in creating such a tree.
Lack of information – As much as we know about the world and its vast number of organisms, we don’t know nearly enough. New species are discovered all the time. New fossil finds occur frequently. New functions for DNA sequences are being uncovered. The list goes on and on. What does this mean for a TOL? It shows that we are nowhere near knowing enough to fully understand life’s complexity. Often, scientific ignorance such as this is exploited by ID proponents to allow for a god of the gaps argument.
Imperfect models – As with any other mathematical model used to predict the world, the algorithms used to create phylogenetic trees are imperfect. In fact, with the exact same dataset, different models will yield different trees. The differences are slight but real. Think about the models that are used to predict hurricane paths. Remember those different colored lines all showing slightly different paths? They all start with the same data, but the models all yield different results. The same issues are present in phylogenetic tree creation.
Confounding biology – The real world is never as simple as we want it to be. In creating phylogenetic trees, we have to contend with biological phenomena that confuse and distort our understanding of evolutionary relationships. This includes such factors as horizontal gene transfer, gene duplication, cross-breading, etc. These factors lead to the crisscrossed and tangled branches that we now know make up TOL.
Gene evolution and speciation are independent – This factor is a subtle, yet important distinction. Speciation can occur instantly (from a geological event or even a single mutation). The accumulation of mutation associated with speciation events could then lag behind what would normally be predicted from speciation.
I hope that I have shed some light on the validity of phylogenetic trees in spite of the numerous problems in creating them. For Part 2, I will directly address Casey Luskin’s points from the Discovery Institute’s Evolution News and Views blog.
Creation of the phylogenetic tree
First, I downloaded the sequences for the cytochrome B genes from the listed species. I picked cytochrome B because I could find it in all the species. The sequences were found on the NCBI website (shown below in small font). After obtaining all the sequences, I entered them into a program called Clustal W. This program aligns DNA or protein sequences. I chose the Kyoto University Bioinformatics Center because it has a nice interface. After aligning my sequences, I chose the option to create a tree based on the N-J algorithm. The result is the tree above.
>gorilla (gorilla gorilla)
ATGACCCCTATACGCAAAACTAACCCACTAGCAAAACTAATTAACCACTCATTCATTGACCTCCCTACCCCGTCCAACATCTCCACATGATGAAACTTCGGCTCACTCCTTGGTGCCTGCTTAATCCTTCAAATCACCACAGGGCTATTCCTAGCCATACACTACTCACCTGATGCCTCAACCGCCTTCTCATCAATCGCCCACATCACCCGAGATGTAAACTATGGCTGAACCATCCGCTACCTCCACGCTAACGGCGCCTCAATATTCTTCATTTGCCTCTTTCTACACATCGGCCGGGGCCTATACTACGGCTCATTTCTCCACCAAGAAACCTGAAACATCGGCATCATCCTCCTACTCACAACCATAGCAACAGCCTTCATAGGCTATGTCCTCCCATGAGGCCAAATATCCTTCTGAGGGGCCACAGTAATCACAAACTTGCTATCCGCCATCCCGTACATCGGAACAGATCTAGTCCAATGAGTTTGAGGTGGTTACTCAGTAGATAGCCCTACCCTTACACGATTCTTTACCTTCCACTTTATCCTACCCTTCATTATCACAGCCCTAACAACCCTCCATCTCCTATTTCTACACGAAACAGGATCAAACAACCCTCTAGGCATCCCCTCCCACTCTGACAAAATCACCTTCCACCCCTACTACACAATCAAAGACATCCTAGGCCTATTCTTCTTTCTCCTGACCTTGATAACATTAACACTATTCTCACCAGACCTCCTAGGAGACCCAGACAACTACACTTTAGCCAACCCCCTAAACACCCCACCCCACATCAAACCCGAATGATATTTCCTATTTGCCTACGCAATTCTCCGATCTGTCCCCAATAAACTAGGAGGCGTCTTAGCTCTATTACTATCCATTCTCATCCTAACAATAATTCCTATTCTCCACATATCCAAACAACAAAGCATAATATTCCGCCCATTAAGCCAACTACTCTACTGATTCCTAATCGCAAACCTCTTCACCCTAACCTGAATCGGAGGACAACCAGTAAGCTACCCCTTCATTACCATTGGGCAAGTAGCATCCGTACTATACTTCACGACAATCCTATTCCTGATACCAATCACATCCCTGATCGAAAACAAAATACTCAAATGAACCT
>orangutan (Pongo abelii)
ATGACCTCAACACGTAAAACCAACCCACTAATAAAATTAATCAACCACTCACTTATCGACCTCCCCACCCCATCAAACATCTCCGCATGATGGAACTTCGGCTCACTCCTAGGCGCCTGCTTAATCATCCAAATCACCACTGGACTATTCCTAGCTATACATTATTCACCAGACGCCTCCACTGCCTTTTCATCAATCGCCCACATCACTCGAGATGTAAACTACGGCTGAATAATTCGCCACCTCCACGCTAACGGCGCCTCAATATTCTTTATCTGCCTCTTCTTACATATCGGCCGAGGCCTATACTATGGCTCATTCACCCACCTAGAAACCTGAAACATCGGCATCATCCTACTATTTACAACTATAATAACAGCCTTCATAGGTTACGTCCTCCCATGAGGCCAAATATCCTTCTGAGGAGCCACAGTAATCACAAATCTACTGTCCGCCATCCCATACATTGGAACAGACCTGGTCCAATGAGTCTGAGGTGGCTACTCAGTAAATAGCCCCACTCTAACACGATTCTTCACCCTACACTTCATACTACCCTTCATTATTACAGCCCTAACAACTCTACACCTCTTATTCCTACACGAAACAGGATCAAATAACCCCCTGGGAATCCCCTCCCATTCCGACAAAATCACCTTCCACCCCTACTACACAATCAAAGACATCCTAGGCCTACTCCTTTTTCTCCTCGCCCTAATAACACTAACACTACTCTCACCAGACCTCCTAAGCGACCCAGACAACTACACCTTAGCTAACCCCCTAAGCACCCCACCCCACATTAAACCCGAATGATATTTCCTATTCGCCTACGCAATCCTACGATCCGTCCCCAACAAACTAGGAGGTGTAATAGCCCTCATACTATCCATCCTAATCCTAACAACAATCCCTGCCCTTCACATGTCCAAGCAACAGAGCATAACATTTCGCCCATTGAGCCAATTCCTATATTGACTTTTAATCGCCGACCTTCTAATTCTCACCTGAATTGGAGGGCAACCAGTAAGCTACCCCTTCATCACCATTAGCCAAGTAGCATCCACATTGTACTTCACTACTATCCTTCTACTTATACCAGCCTCTTCCCTGATCGAAAACCACATACTCAAATGAACCT
>human (homo sapiens)
ATGACCCCAATACGCAAAATTAACCCCCTAATAAAATTAATTAACCACTCATTCATCGACCTCCCCACCCCATCCAACATCTCCGCATGATGAAACTTCGGCTCACTCCTTGGCGCCTGCCTGATCCTCCAAATCACCACAGGACTATTCCTAGCCATACACTACTCACCAGACGCCTCAACCGCCTTTTCATCAATCGCCCACATCACTCGAGACGTAAATTATGGCTGAATCATCCGCTACCTTCACGCCAATGGCGCCTCAATATTCTTTATCTGCCTCTTCCTACACATCGGGCGAGGCCTATATTACGGATCATTTCTCTACTCAGAAACCTGAAACATCGGCATTATCCTCCTGCTTGCAACTATAGCAACAGCCTTCATAGGCTATGTCCTCCCGTGAGGCCAAATATCATTCTGAGGGGCCACAGTAATTACAAACTTACTATCCGCCATCCCATACATTGGGACAGACCTAGTTCAATGAATCTGAGGAGGCTACTCAGTAGACAGTCCCACCCTCACACGATTCTTTACCTTTCACTTCATCTTACCCTTCATTATTGCAGCCCTAGCAGCACTCCACCTCCTATTCTTGCACGAAACGGGATCAAACAACCCCCTAGGAATCACCTCCCATTCCGATAAAATCACCTTCCACCCTTACTACACAATCAAAGACGCCCTCGGCTTACTTCTCTTCCTTCTCTCCTTAATGACATTAACACTATTCTCACCAGACCTCCTAGGCGACCCAGACAATTATACCCTAGCCAACCCCTTAAACACCCCTCCCCACATCAAGCCCGAATGATATTTCCTATTCGCCTACACAATTCTCCGATCCGTCCCTAACAAACTAGGAGGCGTCCTTGCCCTATTACTATCCATCCTCATCCTAGCAATAATCCCCATCCTCCATATATCCAAACAACAAAGCATAATATTTCGCCCACTAAGCCAATCACTTTATTGACTCCTAGCCGCAGACCTCCTCATTCTAACCTGAATCGGAGGACAACCAGTAAGCTACCCTTTTACCATCATTGGACAAGTAGCATCCGTACTATACTTCACAACAATCCTAATCCTAATACCAACTATCTCCCTAATTGAAAACAAAATACTCAAAT
>Chimpanzee (Pan troglodytes)
ATGACCCCGACACGCAAAATTAACCCACTAATAAAATTAATTAATCACTCATTTATCGACCTCCCCACCCCATCCAACATTTCCGCATGATGGAACTTCGGCTCACTTCTCGGCGCCTGCCTAATCCTTCAAATTACCACAGGATTATTCCTAGCTATACACTACTCACCAGACGCCTCAACCGCCTTCTCGTCGATCGCCCACATCACCCGAGACGTAAACTATGGTTGGATCATCCGCTACCTCCACGCTAACGGCGCCTCAATATTTTTTATCTGCCTCTTCCTACACATCGGCCGAGGTCTATATTACGGCTCATTTCTCTACCTAGAAACCTGAAACATTGGCATTATCCTCTTGCTCACAACCATAGCAACAGCCTTTATGGGCTATGTCCTCCCATGAGGCCAAATATCCTTCTGAGGAGCCACAGTAATTACAAACCTACTGTCCGCTATCCCATACATCGGAACAGACCTGGTCCAGTGAGTCTGAGGAGGCTACTCAGTAGACAGCCCTACCCTTACACGATTCTTCACCTTCCACTTTATCTTACCCTTCATCATCACAGCCCTAACAACACTTCATCTCCTATTCTTACACGAAACAGGATCAAATAACCCCCTAGGAATCACCTCCCACTCCGACAAAATTACCTTCCACCCCTACTACACAATCAAAGATATCCTTGGCTTATTCCTTTTCCTCCTTATCCTAATGACATTAACACTATTCTCACCAGGCCTCCTAGGCGATCCAGACAACTATACCCTAGCTAACCCCCTAAACACCCCACCCCACATTAAACCCGAGTGATACTTTCTATTTGCCTACACAATCCTCCGATCCATCCCCAACAAACTAGGAGGCGTCCTCGCCCTACTACTATCTATCCTAATCCTAACAGCAATCCCTGTCCTCCACACATCCAAACAACAAAGCATAATATTTCGCCCACTAAGCCAACTGCTTTACTGACTCCTAGCCACAGACCTCCTCATCCTAACCTGAATCGGAGGACAACCAGTAAGCTACCCCTTCATCACCATCGGACAAATAGCATCCGTATTATACTTCACAACAATCCTAATCCTAATACCAATCGCCTCTCTAATCGAAAACAAAATACTTGAATGAACCT
>Gibbon (Hylobates lar)
ATGACCCCCCTGCGCAAAACTAACCCACTAATAAAACTAATCAACCACTCACTTATCGACCTTCCAGCCCCATCCAACATTTCTATATGATGAAACTTTGGTTCACTCCTAGGCGCCTGCTTGATCCTCCAGATCATCACAGGATTATTTTTAGCCATACACTACACACCAGATGCCTCCACAGCTTTCTCATCAGTAGCTCACATCACCCGAGACGTAAACTACGGCTGAATCATCCGCTACCTTCACGCCAACGGTGCCTCAATATTTTTTATCTGCCTATTCCTACACATCGGCCGAGGCCTATACTACGGTTCATTCCTTTACCTAGAAACCTGAAATATTGGCATTATCCTCCTACTCGCAACCATAGCAACAGCCTTCATGGGCTATGTCCTCCCATGAGGCCAAATATCCTTTTGAGGGGCCACAGTAATCACAAACCTACTATCCGCCGTCCCATACATCGGAACAGATCTAGTCCAATGGGTCTGAGGCGGCTACTCAGTAGATAACGCCACACTCACACGCTTTTTCACCTTTCACTTCATCCTACCTTTCATTATCACGGCCCTAGCAGCCCTGCACCTTCTATTCCTACACGAGACAGGATCAAACAATCCCTTAGGCATCTCCTCCCAACCAGACAAAATCGCCTTCCACCCCTACTATACAATCAAAGACATCCTAGGACTATTTCTCCTCCTCCTCATACTAATAAGCCTAGTACTATTCTCACCCGACCTCCTAGGCGACCCGAGCAACTATACCCAGGCTAATCCCCTAAACACCCCTCCCCACATCAAACCCGAATGATACTTTTTATTCGCATACGCAATTCTACGGTCCGTCCCTAATAAATTGGGAGGCGTACTAGCCCTCCTACTATCAATCCTCATCCTAGCAATAATCCCCGCACTCCACACAGCTAAACAGCAAAGCATGATATTTCGCCCACTAAGCCAGCTCACGTACTGACTCCTAGTAATAAACTTACTGATTCTCACATGAATCGGAGGACAACCGGTAAGCTACCCATTTATCACCATTGGACAAGTGGCATCCGCACTATACTTCACCACAATCCTAGTACTTATACCAGCCGCCTCCCTAATCGAAAACAAAATACTCAAATGAACCT
>Rhesus Macaque (Macaca mulatta)
ATGACTCCAATACGCAAATCCAACCCAATCCTAAAAATAATTAATCGCTCCTTCATCGATTTACCCGCCCCACCCAACCTCTCCATATGGTGAAACTTTGGCTCACTTCTTGCAGCCTGCCTAATTTTACAAATCATCACAGGCCTACTCCTAGCAATACACTACTCACCAGACACCTCCTCCGCCTTCTCCTCAATTGCACATATCACCCGAGATGTAAAGTACGGCTGAATCACTCGCTACCTCCACGCCAATGGTGCCTCTATACTCTTCATTTGCCTTTTCCTACACATCGGTCGGGGCCTTTACTACGGCTCATACCTCCTCCTAGAAACCTGAAACATTGGTATTATACTCCTTCTTATAACTATAACAACGGCTTTCATGGGTTATGTTCTCCCATGAGGCCAAATATCATTCTGGGGAGCAACAGTAATCACAAACCTGCTATCAGCAATCCCGTATATCGGAACCAATCTCGTCCAATGAATCTGAGGAGGATACGCCATCGACAGCCCTACTCTCACACGATTCTTCACCTTACACTTTATCCTACCCTTCATCATCATCGCCCTCACAACCGTGCACCTACTATTCCTGCACGAAACAGGATCAAACAACCCTTGCGGAATCTCCTCCGACTCAGACAAAATCGCCTTCCACCCCTACTACACAACCAAAGACATCCTGGGCCTAGTCCTCCTTCTCTTCATCCTAGCAACACTAACACTACTCTCACCCAACCTCCTAAACGACCCAGACAACTACATTCCAGCCGACCCATTAAACACTCCCCCACATATCAAACCAGAGTGATACTTCCTATTTGCATACACAATCCTACGATCCATCCCCAACAAACTGGGAGGCGTACTAGCACTCTTTCTATCGATCCTCATTCTAGCAGCCATCCCTATACTTCACAAATCCAAACAACAAAGCATAATATTCCGCCCACTCAGCCAATTTCTATTCTGACTCCTAATCACAATCCTATTGACCCTTACCTGAATTGGAAGCGAACCAGTAGTCCAACCCCTTACCACTATCGGCCAAGTAGCATCCATAATATACTTCATCACAATTCTAATCCTAATACCACTGGCCTCCCTAATCGAAAACAACCTACTCAAGTGAACTT
>White-fronted Capuchin (Cebus albifrons)
ATGACCTCTCCCCGCAAAACACACCCATTAATAAAAATTATTAATAGTTCATTTATTGATCTGCCCACACCATCCAACATCTCCTCCTGATGAAACTTCGGATCACTTCTAGGCGCCTGCCTAATAATTCAAATTACCACAGGCCTATTCTTAGCGATACACTATACGCCAGACACCTCAACCGCCTTCTCCTCAGTAGCACATATTACCCGAGATATTAATTACGGTTGAATAATCCGCCTCCTACACGCCAATGGTGCCTCCATATTTTTTGTGTGCTTATTTCTCCACACTGGCCGAGGCCTCTACTACGGATCTTTTCTCTTTCTAAACACCTGAAATATTGGTACAATCCTATTATTAATAACAATAGCCACAGCCTTTATAGGCTATGTCTTACCGTGAGGCCAAATATCATTCTGAGGAGCCACAGTTATTACAAATCTTCTATCAGCCATCCCCTATACCGGACATAACCTTGTACAATGAATCTGAGGTGGCTTTTCAGTAGATAAACCCACCCTCACACGATTCTTTACCTTTCACTTTATTTTACCTTTTATTATCACAGCCCTAGCAACTATTCACCTTTTATTTCTACATGAAACAGGCTCAAATAATCCATCAGGAATAGCATCTAGCCCCGATAAAATTATATTCCATCCCTACTACACAACCAAAGATATTTTTGGATTAACCCTTCTTCTCCTACTCCTTACAAGCCTAACCCTATTTACCCCCGACCTTTTAACTGACCCAGATAACTACACACTAGCCAACCCCCTTAATACTCCACCCCATATTAAGCCAGAGTGATACTTTCTATTCGCATACACAATTTTACGATCTATTCCAAATAAACTAGGAGGTGTTCTAGCTCTTCTATTATCTATTATAATCCTAACAATTATCCCTGCCACTCACCTATCCAAACAACAAAGTATAATATTCCGACCAATCACCCAAATCCTATTCTGAACCCTAGCAGCCGATCTACTTACACTTACATGAATTGGAGGCCAACCAGTAGAATACCCCTTTGAAGCCATTGGCCAAACCGCATCTATTGCTTACTTCCTTATTATTACTCTAATTCCTCTATCAGCCCTAACTGAAAATAAGCTACTTAAATGATAA
Two different techniques yielded the exact same tree. Both trees created here match to a tree that is created by looking at physical similarities and differences. This one example should be enough to show the validity of phylogenetic trees, but you know nothing would be sufficient to convince the Discovery Institute.
So I have shown a couple reasons why I think phylogenetic trees are valid. Keep these in mind when you read about the supposed failures of phylogenetic trees from the ID proponents. Next, I am going to describes some of the difficulties in creating a complete TOL. Remember, these issues do not mean that common descent is wrong, it just means that our ability to decipher the tree of life is limited. A weakness in data is not a weakness in a theory.
Difficulties in creating a tree of life
There are many problems with creating a complete tree of life that takes into account every piece of data available. Think about all the information and computation required for the creation of an all-encompassing phylogenetic tree. In addition, a TOL has to determine the relatedness of organisms that might not have a common ancestor for over half a billion years. Below is an overview of some other problems in creating such a tree.
Filed under: Evidence | Tagged: Evidence, phylogenetic tree | 1 Comment »


